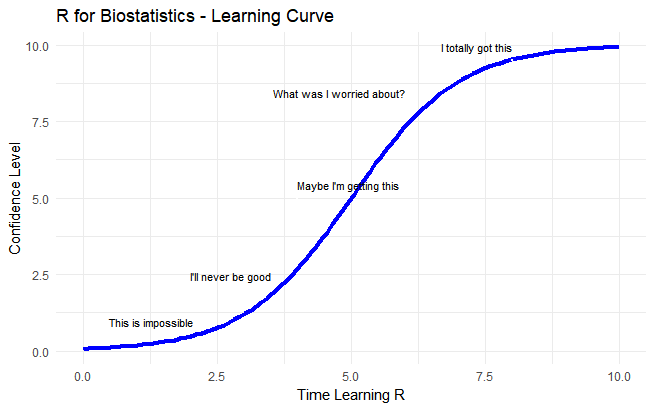
So, you’ve started an MS and want to learn R for biostatistics? Exciting times! But if you’re struggling, you’re not alone.
Many students really struggle to get to grips with R during their Biostatistics MS (about 70% of my masters cohort).
This is a big problem!
A large proportion of Biostatistician jobs will require you to be proficient in R, so if you don’t know the difference between a vector and a DataFrame- you’re going to be limiting your chances in the job market.
In this post I’ll talk about why learning R can feel so difficult and how you can master it effectively.
Why Are You Struggling To Learn R For Biostatistics?
You’re struggling to learn R in your biostatistics MS for two reasons:
a) You’re book-smart, but not as practically inclined.
- You have strong theoretical background but haven’t had much hands-on coding experience.
b) Your university teaches R like theory course.
- Endless PDFs, dry lectures and minimal practical application.
But biostatistics is a highly practical field, you need hands-on experience with R is key to build confidence and competence. You won’t get that if you’re learning R the wrong way!
My Experience Learning R For Biostatistics
I made many mistakes when I first tried to learn R. Here are just some of them:
- Reading textbooks about R instead of coding in R
- Trying to memorise code instead of understanding how it works
- Copy and pasting code from others without grasping the logic
- Jumping into projects before learning the basics
Looking back, I wish I could have saved hours of wasted time by learning R differently.
I can’t go back, but at least I can help you avoid my mistakes and learn R the right way.
How To Learn R For Biostatistics (The Right Way)
In order to learn R for biostatistics applications – you need to learn it in the right order.
The steps you need to take to learn R are as follows:
- Learn syntax and basics
- Solve basic problems
- Follow along with real projects
- Start your own project
You might want to rush these steps or skip a step. This will only lead to frustration!
It’s like learning any language—if you try to write complex sentences before you know the basic words, you’ll struggle.
Similarly, in R, if you try to jump straight into complex modeling without understanding the basics of data manipulation, you’ll likely feel lost.
Step 1: Learn Syntax and Basics
Before you jump into advanced techniques, make sure you understand the syntax and basics.
This includes topics such as variables, data structures, for loops and functions.
How to do this:
- Read or watch a tutorial, but don’t passively consume it – code along in your R console.
- Focus on understanding, not memorizing. In the real world, no one is expecting you to remember everything.
Once you understand the code, move on to the next lesson.
Here are some free resources for you to follow:
- R 4 Biostats (Lab 1)
- The Epidemiologist R Handbook (Basics & Data Management sections)
- FastR (Part I)
- Swirl (this is an R package that guides you through learning R in the R console)
This step may sometimes feel a bit frustrating, but it’s essential.
Before moving to the next step, make sure that you’ve covered all the concepts in one of the above resources.
Step 2: Solve Basic Problems
Now you’ve got to grips with the basics, it’s time for you to apply what you’ve learned.
You’re not quite ready to write your own project yet, but what you can do is solve basic problems.
I recommend you complete a worksheet problems or lab exercises.
Why is this the best way to practice?
- Completing exercises will give you a chance to put your new found R skills into action.
- Comparing your code against the solutions will provide you with immediate feedback, which will help you improve.
- Solving small exercises gives you a sense of accomplishment that you’re actually learning R.
This step can be daunting – you may feel you don’t know enough or you will get stuck.
Don’t worry! There will be exercises where you haven’t covered the background yet. This is by design – this is what real life biostatisticians encounter all the time.
If you get stuck – go back to the resources above and look for answers, google to help you remember syntax or data types.
Remember, this isn’t cheating – this is exactly what many biostatisticians do daily in our jobs to help rejog our memory.
One thing I don’t recommend you doing is to find the answer using google or chatGPT (it can be wrong). You won’t be learning – you will be copying.
You can use these resources to answer questions like “how do I run a for loop in R” but not “A scientist needs to experiment upon 4 conditions, 5 times each. Generate a vector of length 20, representing these conditions in R.”
Notice how the first gives you tools to solve the problem, the second answers the problem for you! Here are a some free exercise worksheets with solutions:
- R Handbook for Biostatistics – Focused on plant biology but the exercises hold up (CTRL+F “Exercise” to find exercises)
- W3Resource R Programming Exercises
- Intro to R (Search for “Exercise” to find exercises)
If you’ve managed to answer all the questions in one of the worksheets – feel free to move onto the next step.
Step 3: Follow Along with Real Projects
Now that you’ve built a foundation, it’s time to see how R is used in real biostatistics projects.
Finding industry projects or someone’s PhD code might prove difficult at this step but you can still follow along with real projects.
The best way to find these projects is with kaggle.
Kaggle is a community for biostatisticians and data scientists to share, test, and stay up-to-date on all the newest techniques. There is a huge repository of community-published projects. The best thing is that they are awarded with upvotes if they are useful. This is where you’re going to discover a project to follow along with.
Your goal:
- Read through someone else’s project code.
- Try to understand their code – don’t just skim it.
- As yourself questions – what is the code doing? Why have they used a specific function?
ChatGPT can be used at this step to clarify anything you don’t understand – but try to understand it yourself first.
Here are some of the best biostatistics projects in R to follow along with:
If you’re comfortable with what the code is doing, move on to the next step.
Step 4: Start Your Own Project
Now for the exciting part – to start your own project.
To start, you can try to run a similar analysis from the project in the last section but run it for a different dataset.
Don’t look at the analysis others have done on the dataset you’ve chosen to avoid any preconceptions about what analysis to do or what features the dataset may have.
If you feel confident enough, feel free to design the analysis yourself. Here are some sections you can use to guide you in your analysis:
- Exploratory Data Analysis (EDA)
- Data Cleaning (if needed)
- Data Transformation
- Model Building
- Model Evaluation
- Model Interpretation
- Model Visualization
If you get stuck – the R and Biostatistics communities are incredibly active and willing to help. Don’t hesitate to ask questions on Stack Overflow and community subreddits (r/biostatistics, r/RLanguage, r/rstats).
Here are some biostatistics datasets you can use:
Conclusion
Follow these steps to learn R for biostatistics! By following these steps, you’ll not only improve your skills for your MS but also enhance your job prospects by building strong technical skills that employers value (remember to add the project to your resume).
Ready to Land More Biostatistics Interviews?
Get my free email course that walks you through the exact steps biostatistics professionals are using to get more callbacks, without rewriting their resume 100 times.
- 🔬 Step-by-step, field-tested advice
Tried-and-true strategies designed specifically for biostatistics job seekers. - ✅ Actionable steps you can apply immediately
Each email includes one clear step to help you get noticed and get interviews. - 📬 1 email per day. Zero fluff. Just results.
Short, focused lessons that respect your time—and deliver real value.